What is oga.jod.li?
The https://oga.jod.li site is a way to distribute the evolution of genetic algorithm with the ability to spread the population through niche mechanisms to avoid local optima and the possibility to improve the speed of the evolution through auto optimisation of the genetic algorithm parameters (mutation, crossover, elitism, niche, chunks) through another genetic algorithm.
As shown below, the client device retrieves chromosomes to evaluate from the oga.jod.li server and sends back the fitness for the received chromosomes.
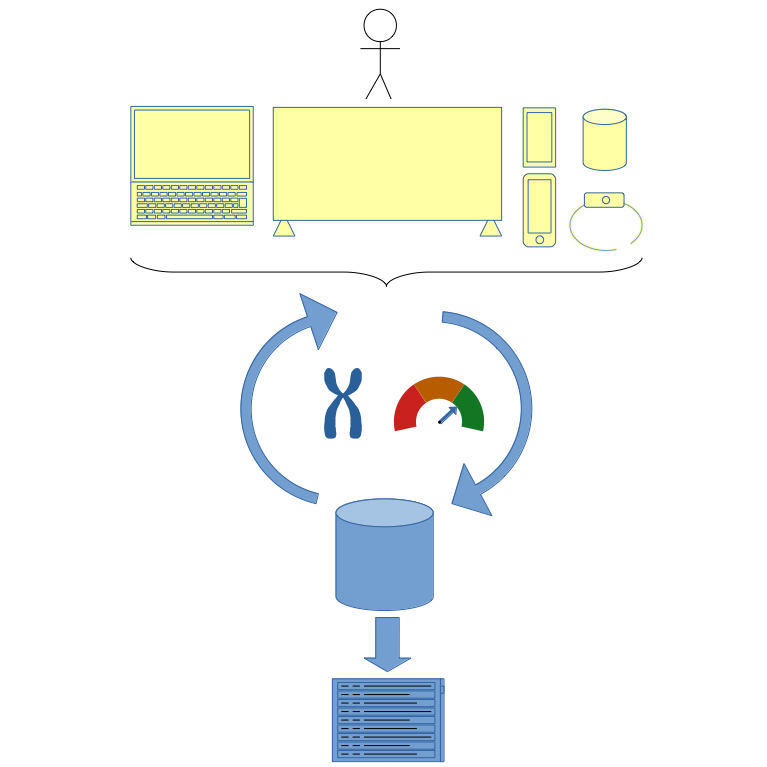
The devices that can evaluate the chromosome fitness can be very diverse:
- computer
- smart TV
- smartwatch
- smartphone
- connected objet
- …
The only requirement is the ability to call the oga.jod.li urls to get chromosomes for evaluation from oga.jod.li server and to send the evaluated chromosome fitness back to the oga.jod.li server.
Why did you start this initiative?
We had the need for a simple distributed genetic algorithm that would be accessible from all sort of devices. So, we built oga.jod.li and decided we can share the solution with others.
Which are the main features of oga.jod.li?
| Feature | Accessible through oga.jod.li site | Accessible through url call | More information |
|---|---|---|---|
| User creation | X | Through email or OAuth2 validation (StackExchange, Google, GitHub). Mandatory to use oga.jod.li . | |
| Population creation/edition | X | X | Mandatory to use oga.jod.li . See doc below. |
| View list of populations | X | Displays the performance, selects population for urls updates/parameters . | |
| Plots | X | Shows the performance (min, max, average) and the evolving parameters of the genetic algorithm population batches. See doc below. | |
| Get chromosomes to evaluate | X | Mandatory to use oga.jod.li . See doc below. | |
| Send evaluated chromosome fitness | X | Mandatory to use oga.jod.li . See doc below. | |
| Migrate n best chromosomes from another population | X | X | See doc below. |
| Get all evaluated chromosomes | X | X | See doc below. |
| Delete old evaluated chromosomes | X | X | See doc below. |
| Renew user key | X | For security reasons, one can renew the key to ensure the account is not compromised. | |
| Delete user | X | Once a user is deleted, all the data related to this user (chromosomes, populations, batches, …) is deleted as well. | |
| Protection against DDoS and brute force attacks | X | X | oga.jod.li is protected against DDOS and brute force attacks so that keys are not guessed and functionalities remain available. |
| Forgotten user or key | X | A user who created an account through email can receive the user name and key details by email. |
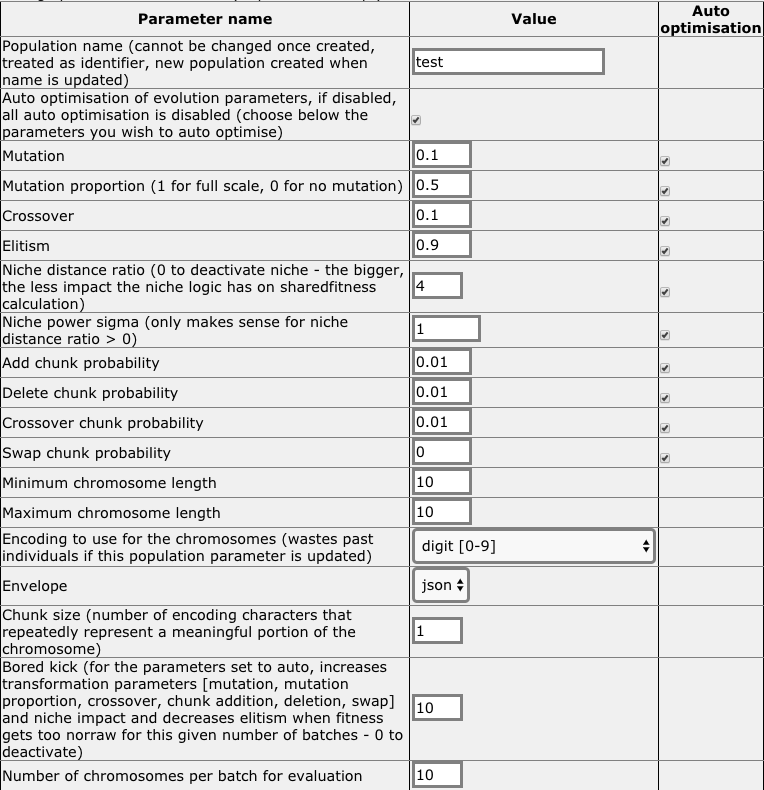
How to create or edit a population?
The genetic algorithm population is made of parameters which are defined as follows:
Population name
The population name which must be unique per user. If the population name doesn’t already exist, the server creates a new population with that name. If the population name already exists for your user, the server updates the population.
Auto optimisation
The auto optimisation parameter. If enabled, the 10 parameters that can be optimised (marked with [auto] below) and for which the auto optimisation is enabled will be automatically optimised. The performance of your evolution can be greatly increased with the help of auto optimisation as shown here: oga.jod.li genetic algorithm auto optimisation
Mutation rate
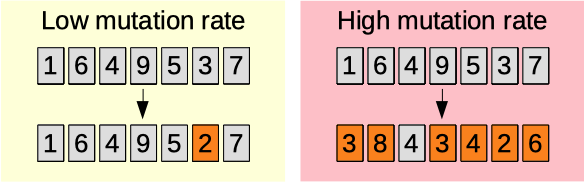
The mutation rate parameter (0.01-0.50) [auto]. The higher, the more the population will be randomly affected. See this link for more information.
Mutation proportion
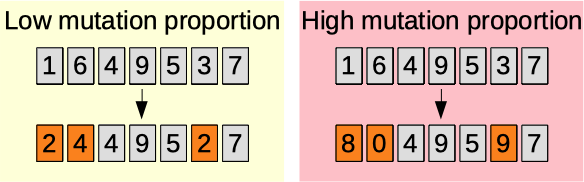
The mutation proportion parameter (0.00-1.00) [auto]. The higher, the more the mutated characters are changed.
Crossover rate
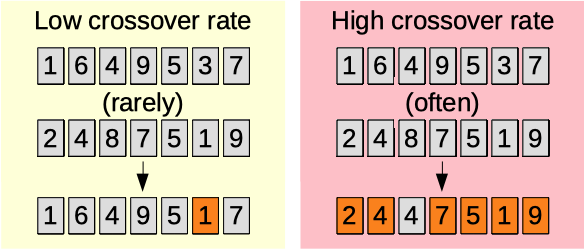
The crossover rate parameter (0.01-0.50) [auto]. The higher, the more a chromosome is mixed with a second parent. More often and with more impact. See this link for more information.
Elitism rate
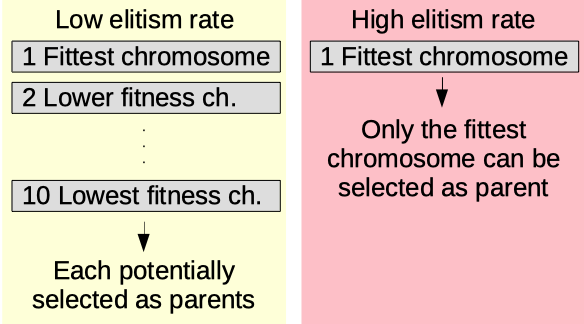
The elitism rate parameter (0.50-1.00) [auto]. The higher, the more likely the fittest chromosomes will be parents of the next ones.
Niche distance ratio
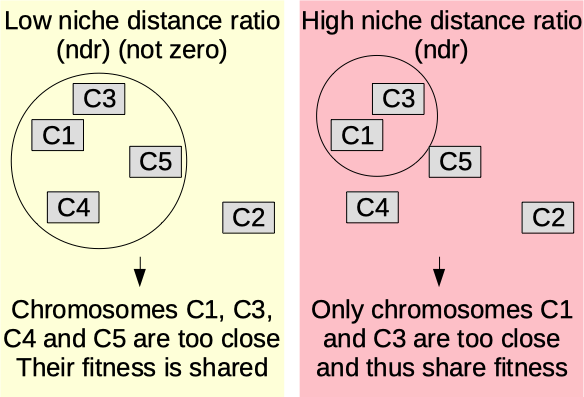
The niche distance ratio (ndr) (0-100) [auto]. Disable niche with 0. Otherwise, higher value means lower penalty on the fitness (shared fitness) for chromosomes being close to each other. More information on the niche mechanism here: oga.jod.li niche mechanism .
Niche power sigma
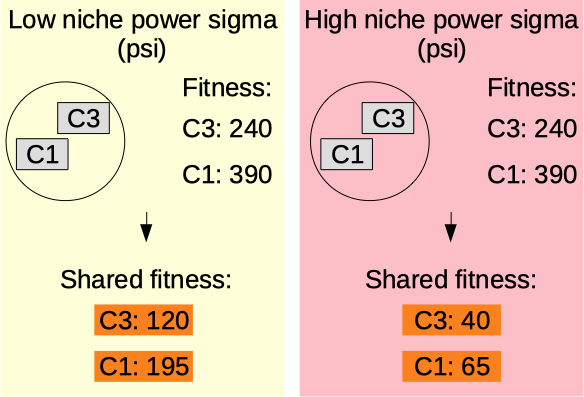
The niche power sigma (psi) (1.00-10.00) [auto]. Only makes sense if ndr != 0. Higher value means higher penalty on the fitness (shared fitness) for chromosomes being close to each other. More information on the niche mechanism here: oga.jod.li niche mechanism .
Chunk size
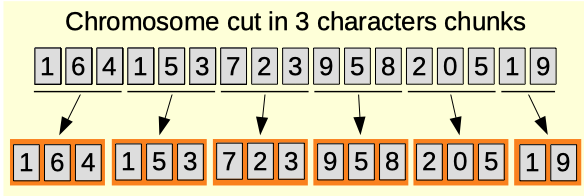
The chunk size parameter (0-100). Minimal number of characters to have a meaningful representation within a chromosome. For example, the chromosomes can represent a lists of colours each encoded as RGB values on 3 bytes. In that case, the chunk size would be 3 with an encoding on xdigit. The chunk size is useful for the 4 below chunk parameters. To disable this and the effects of the chunk parameters, set the chunk size to 0.
Add chunk
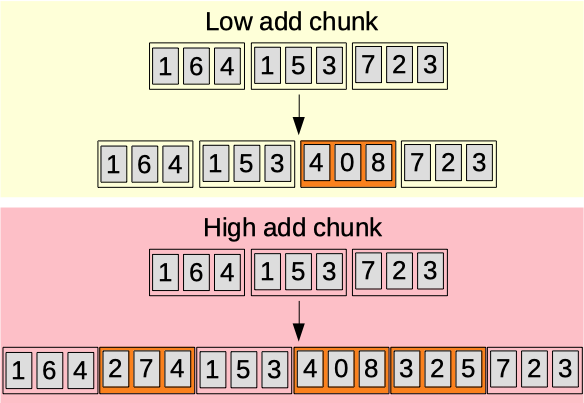
The add chunk parameter (0.00-1.00) [auto]. Sets the probability for a chunk to be inserted between two other chunks.
Delete chunk
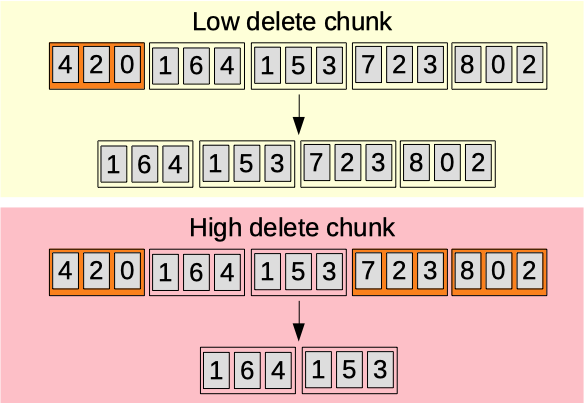
The delete chunk parameter (0.00-1.00) [auto]. Sets the probability for a chunk to be deleted.
Chunk crossover
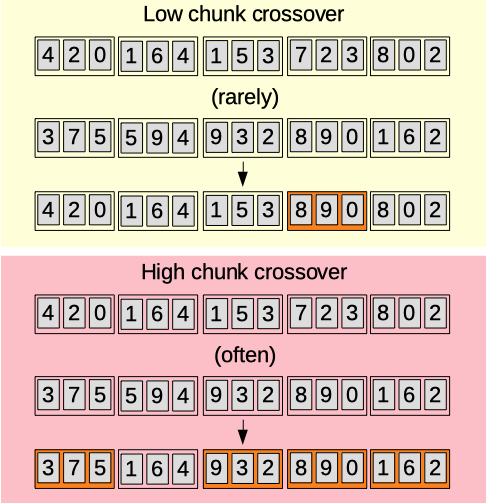
The crossover chunk parameter (0.00-1.00) [auto]. Sets the probability for a chunk to be replaced by another one at the same position from another chromosome.
Swap chunk
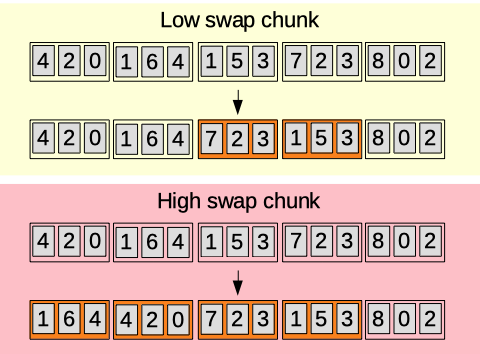
Length
The minimum length of the chromosomes (1-3000).
The maximum length of the chromosomes (1-3000). Must be higher than previous.
Encoding
The encoding of the chromosomes can be:
- alnum [a-zA-Z0-9]
- alpha [a-zA-Z]
- lower [a-z]
- upper [A-Z]
- xdigit [A-F0-9] (hexadecimal)
Envelope
The envelope of the HTTPS responses can be:
- JSON
- XML
- CSV
Chromosomes per batch
The number of chromosomes per batch. Use a small number if you wish to use many devices at the same time. Use a large number if you have a poor connection.
Bored kick
The bored kick parameter is the number of batches to evaluate for a population to be considered boring and deserving a kick. The kick is given to some parameters provided they are set to be auto optimised.
Increase of parameters:
- mutation
- mutation proportion
- chunk addition
- chunk deletion
- chunk swap
Decrease of parameters:
- elitism
- niche distance ratio (depends on the spread of the population)
This applies only if the latest batch does not contain the highest fitness chromosome.
The general outcome regarding the bored kick parameter through experiment results is that it leads to best fitnesses more concentrated as shown here.
Population creation and edition
The population can be created and the parameters can be edited from the https://en.oga.jod.li page or through a url similar to the following one:
https://en.oga.jod.li/?api=pop&u=toto&k=1234567890123456&pop=test&auto=1&mut=0&cro=0.1&eli=0.9&ndr=4&psi=1&min=10&max=10&enc=digit&env=json&nb=10&mp=0.5&ck=1&ac=0.01&dc=0.01&cc=0.01&sc=0&am=1&=1&aac=1&adc=1&acc=1&asc=0&acs=1&ae=1&an=1&ap=1
Where the user toto and the key 1234567890123456 must be replaced by the ones you got on the https://en.oga.jod.li page either through email or through OAuth2 (StackExchange, Google, GitHub) authentication. This url is made available and customised for you on the https://en.oga.jod.li page based on the parameter values you enter:
How to get chromosomes to evaluate?
Request chromosomes to evaluate
The url to get a batch of chromosomes for evaluation contains:
- The server name to get the chromosomes. This will be en.oga.jod.li .
- The user name you created on oga.jod.li .
- The user key. Make sure nobody else gets access to your key.
- The population name for which you wish to evaluate chromosomes.
The url looks like the following:
https://en.oga.jod.li/?api=gc&u=toto&k=1234567890123456C&pop=test
Here, the user is “toto”, the key is “1234567890123456” and the population name is “test”. You can find this url for your user and the selected population on https://en.oga.jod.li .
Response with chromosomes to evaluate
The server response to this call depends on the population parameters. It always contains the chromosome and the chromosome id. Find below few examples with batch size of 3 and chromosome length bound between 1 and 10:
Encoding: digit
{
"pop": "test",
"enc": "digit",
"status": "to evaluate",
"chromosome": [{
"id": "1313193",
"chromosome": "554530"
}, {
"id": "1313194",
"chromosome": "3213529939"
}, {
"id": "1313195",
"chromosome": "101900860"
}]
}
<pop name="test" enc="alnum" status="to evaluate">
<chromosome id="1313177">QBsgo</chromosome>
<chromosome id="1313178">SITP8wDB</chromosome>
<chromosome id="1313179">YA53xfE</chromosome>
</pop>
1313181,377E53C369631840B4
1313182,25FC
1313183,F91E
Any combination of encoding and envelope population parameter is valid.
How to send the chromosome fitness?
Once the chromosome fitness is evaluated on your device, you need to send it to the oga.jod.li server using the following url:
https://en.oga.jod.li/?api=scf&u=toto&k=1234567890123456&id=1234&fitness=2.3
This url is prepared for your user on the https://en.oga.jod.li page. You need to programatically adapt the chromosome id and the fitness values for this url call to make sense.
Distribute the evolution of genetic algorithm
One can distribute the evolution of genetic algorithm with oga.jod.li by requesting chromosomes to evaluate, evaluating their fitness and sending back their fitness from several client devices within the same population. The chromosomes sent to one client device are not sent to another client device. Thus, the client devices do not waste time evaluating the same chromosomes.
How to view populations?
Once chromosomes are being evaluated, one may wish to follow the performance of the population. This is possible from the https://en.oga.jod.li page with the populations selection view:
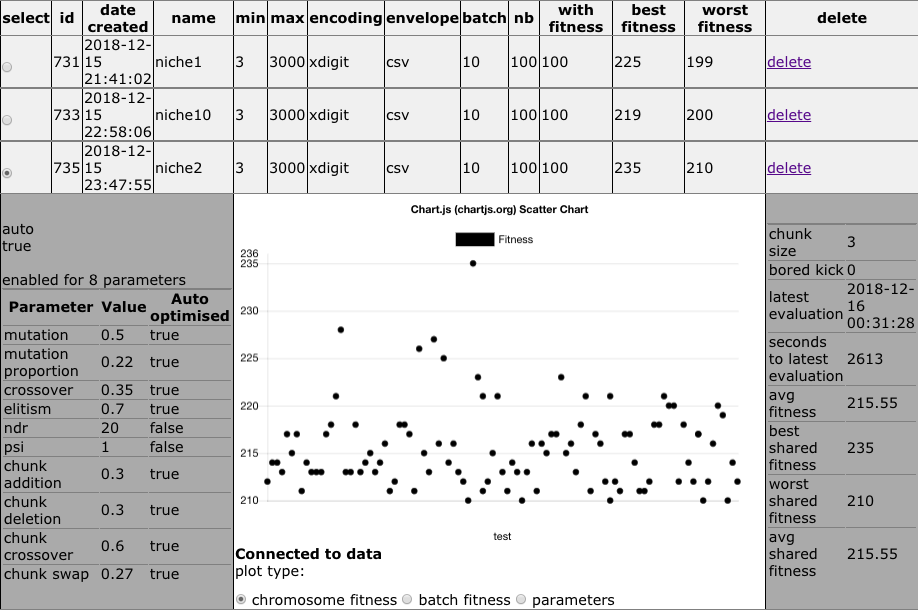
Selecting a population changes the urls of the other features on the page, displays the details and plots for that population.
The plots are built with the help of https://chartjs.org . They grow as the data flows in from the evolution through websockets. They are of the following types:
- Chromosome fitness: follow the fitness of up to 300 evaluated chromosomes.
- Batch fitness: follow the max, min, average fitness of the chromosome batches.
- Batch parameters: follow the genetic algorithms that can be automatically evolved (mutation, crossover, elitism, niche, chunk) for each batch – relevant when the population is set to be auto optimised.
Each population can be deleted from the list of populations. This deletes the population, the chromosomes and the batches. No information related to this population remains on the oga.jod.li server after population deletion.
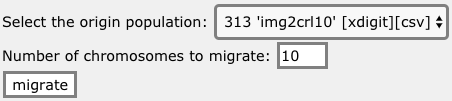
How to migrate the n best chromosomes from one population to another
One way to boost the evolution of a population consists in injecting the best chromosomes from one population to another. To do that, one can use the https://en.oga.jod.li web interface or a url similar to the following on:
https://en.oga.jod.li/?api=mc&u=toto&k=1234567890123456&pop=test&oripop=bettertest&nb=10
This url is prepared for your user, the selected populations and the selected number of chromosomes on the https://en.oga.jod.li page:
How to get all the evaluated chromosomes from one population?
Once the evolution is over, one may want to retrieve the remaining evaluated chromosome (maximum 100). To do that, one can use the https://en.oga.jod.li web interface or a url similar to the following on:
https://en.oga.jod.li/?api=gaec&u=toto&k=1234567890123456&pop=test
This url is prepared for your user and the selected populations on the https://en.oga.jod.li page.
How to delete the old non evaluated chromosomes?
The number of evaluated chromosomes is limited to a maximum of 100 per population. Once this limit is reached, the evaluated chromosomes with the lowest fitness (or shared fitness in case of niche mechanism) are deleted. The maximum number of chromosomes per population is 1000. Once this limit is reached, the oldest non evaluated are deleted if their evaluation takes significantly longer (100 times) than the average time for evaluation within the same population. It can still be that the number of non evaluated chromosomes is too high. For example if there are too many devices on which the evolution is distributed or if the population batch size is too high or when many chromosomes get lost without evaluation. In this case, the oga.jod.li will not send chromosomes for evaluation until some chromosomes are deleted with the following message:
cannot purge enough non evaluated chromosomes - either send evaluations, delete existing chromosomes or wait for the evaluation wait threshold to be reached
To delete old non evaluated chromosomes, one can wait for the oldest chromosomes to be automatically deleted or evaluated or use the https://en.oga.jod.li web interface or a url similar to the following one:
https://en.oga.jod.li/?api=donec&u=toto&k=1234567890123456&pop=test&hours=24
This url is prepared for your user, the selected number of hours and the selected populations on the https://en.oga.jod.li page. The lower the number of hours, the higher the number of chromosomes that could be deleted:

Which programming languages can make use of oga.jod.li genetic algorithm?
Any language that can call a GET url can take part in the genetic algorithm evolution on oga.jod.li . This is true for at least the following:
- JavaScript
- Shell script
- PHP
- Swift
- Java
- C#
- Python
- …
Video
Here is a YouTube video on how to execute an oga.jod.li optimised genetic algorithm for the PNG interlaced file size question as an example on how to use oga.jod.li distributed genetic algorithm with a niche mechanism:
What is next?
Here is a list of features we wish we could implement:
- Upload chromosomes to population.
- Neural network based genetic algorithm parameters optimisation.
- Stop evolution through a button.
- Share a population with another user.
- Share chromosomes with another user.
- Select different types of mutation operators.
- Define allowed character sets.
- Select different niche mechanisms.
- Upload population evolution package that can be downloaded, extracted and executed by client devices.
- Report on client performance for fitness evaluation.
- Apply a fitness factor parameter on migration.
If you are interested in any of those developments or any other related to oga.jod.li, feel free to contact us through the contact form on https://en.oga.jod.li .
